PASSer:
Protein Allosteric Sites Server
Introduction
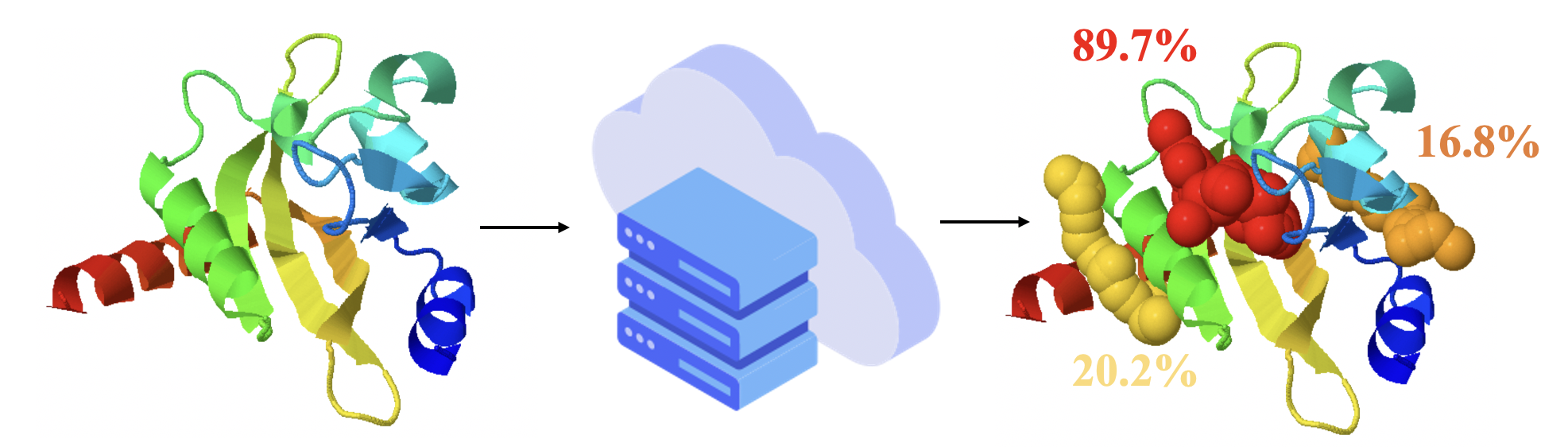
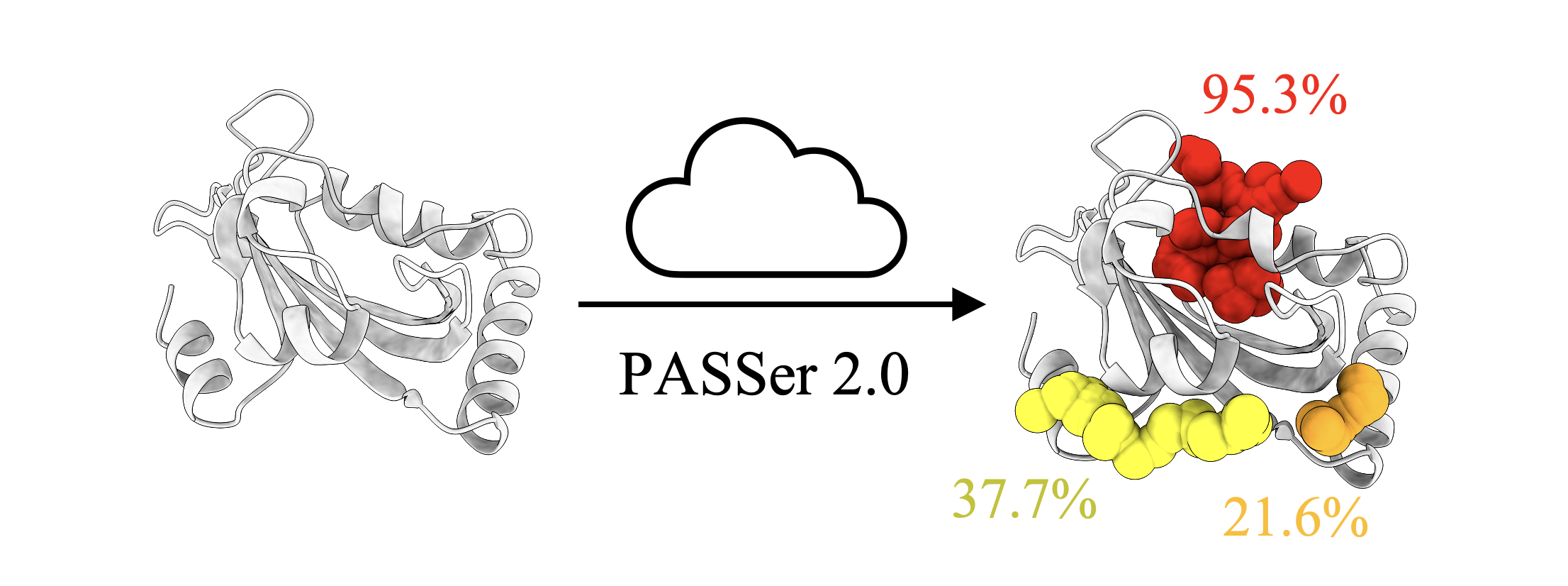
Allostery is the process by which proteins transmit perturbations caused by the binding effect at one site to another distal site. The allosteric process is fundamental in the regulation of activity. Identifying allosteric sites is important for allosteric drug development and has attracted a wide range of interests.
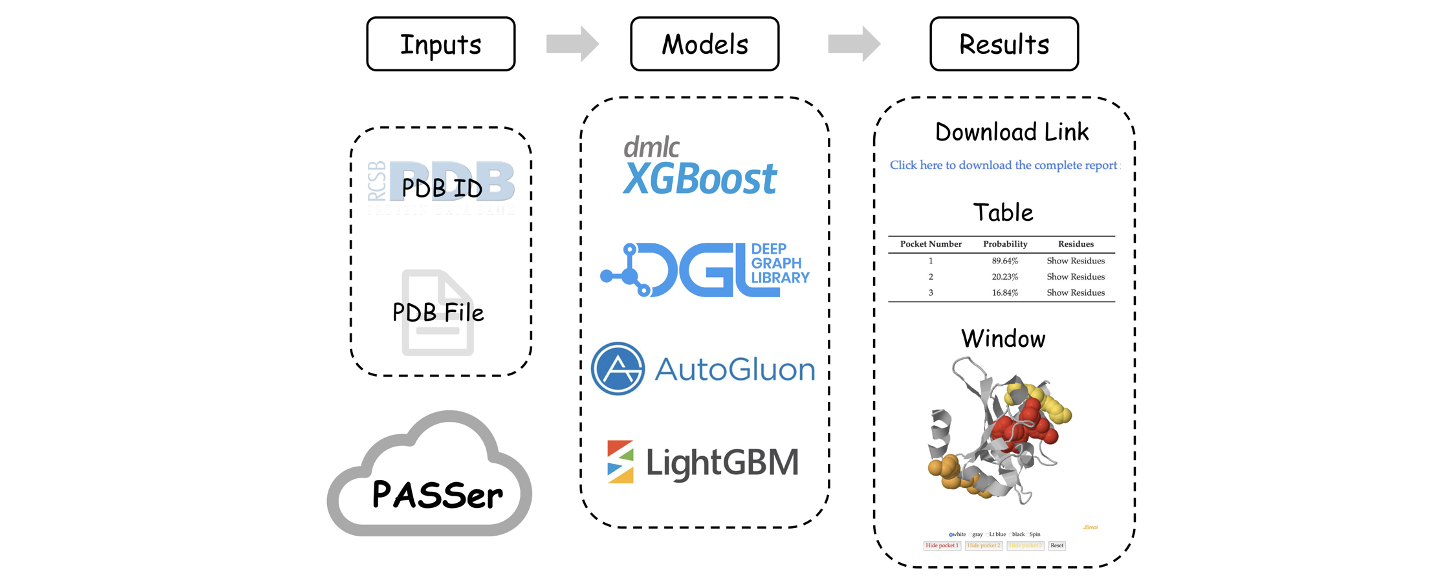
To approach this problem, PASSer is designed for accurate allosteric site prediction. PASSer provides three machine learning models: (1) ensemble learning through extreme gradient boosting and graph convolutional neural network; (2) automated machine learning through AutoGluon framework; and (3) learning to rank through lightgbm.
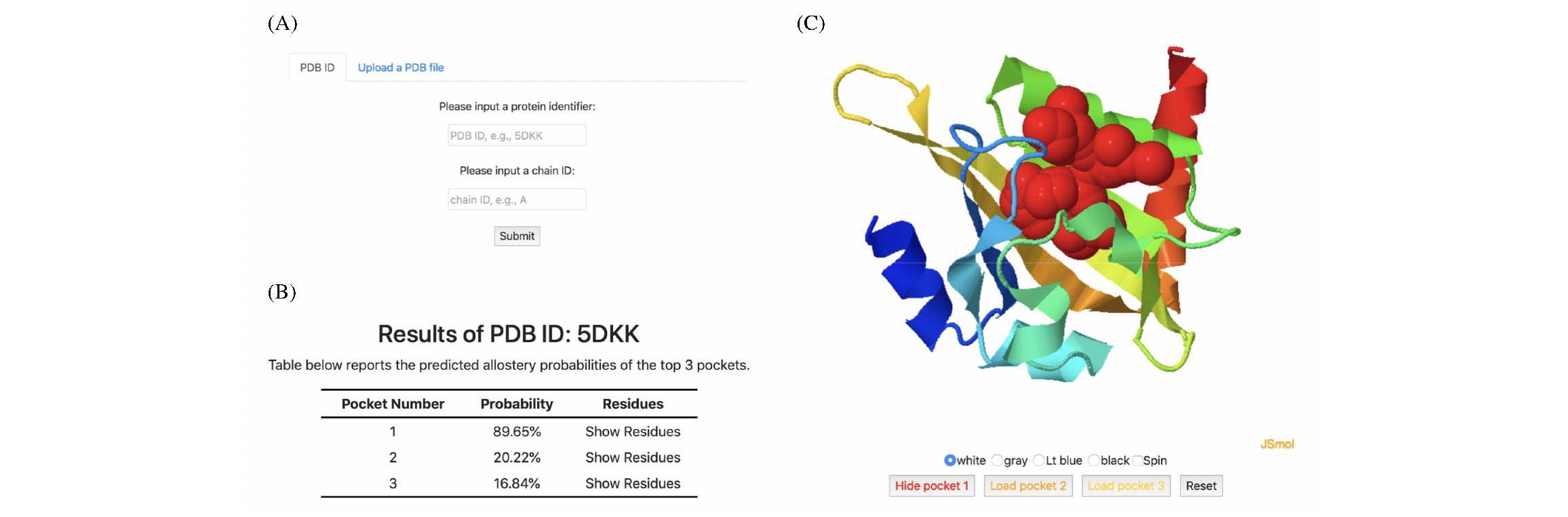
PASSer is deployed with trained machine learning models and has been extensively tested to complete prediction within seconds. The prediction submissions and result retrievals can be accessed via the web page or API.
Note: (1) All uploaded and analyzed data is deleted upon job completion. (2) The website is refreshed every day at 4 AM central time.
Run prediction
DIY
-
Want to use machine learning-ready dataset to train your own model?
- Download it from the server! -
Want to customize the dataset?
- Check our scripts on GitHub to automate your preparation pipeline!